Single pulse analysis¶
Kookaburra provides a command-line tool for fitting single pulses. To get help with this, run
$ kb_single_pulse --help
This takes a single positional argument, the data to analyse, and a number of optional arguments related to the choice of model to fit and priors. We provide a quick-start example on simulated data below.
Quick start¶
To demonstrate the basic input and output, we will create simulated data from
a pulsar (flux as a function of time) and analyse it. First, we run the script
make_fake_data.py available in the examples directory. Here are the
contents:
""" Script to generate a data file with a single simulated pulse
Usage:
$ python make_fake_data.py
This will generate a file fake_data.txt, a comma-separated file of the time
vs. flux for the simulated signal. The simulated signal contains a simple
first-order polynomial base flux and a three-component shapelet pulse.
"""
import numpy as np
import pandas as pd
import kookaburra as kb
# Injection parameters
pulse_injection_parameters = dict(
C0=0.6, C1=0.1, C2=0.2, beta=1e-3, toa=0.005, # Parameters for the shapelets
B0=1, B1=-20 # Parameters for the polynomial base-flux
)
# Instantiate a flux model: a sum of the shaplet and polynomial flux classes
flux_model = kb.flux.ShapeletFlux(3) + kb.flux.PolynomialFlux(2)
# Generate fake data using the instantiated flux model and injection parameters
N = 1000
time = np.linspace(0, 2 * pulse_injection_parameters["toa"], N)
flux = flux_model(time, **pulse_injection_parameters)
# Add Gaussian noise
sigma = 2e-2
flux += np.random.normal(0, sigma, N)
# Write the data to a text file
df = pd.DataFrame(dict(time=time, flux=flux, pulse_number=0))
filename = "fake_data.txt"
df.to_csv(filename, index=False)
This will create a file fake_data.txt.
Now we can run kb_single_pulse on this data file.
$ kb_single_pulse fake_data.txt -p 0 -s 5 -b 2 --plot-fit
The flag -p sets the pulse number to extract from the data file. For
this example simulated data only a pulse 0 exists, but in general a data file
could contain many pulses. The flag -s sets the number of shapelets
to use in the fit. When simulating the data, the model had 3 shapelets. Here,
we set a maximum number of 5. The flag -b sets the order of the base
polynomial. Here we chose a first order polynomial. If you do not need to
remove a baseline, set -b 0. Finally, we include flags to plot the data
(this will be done before the main analysis begins) and the fit to the data.
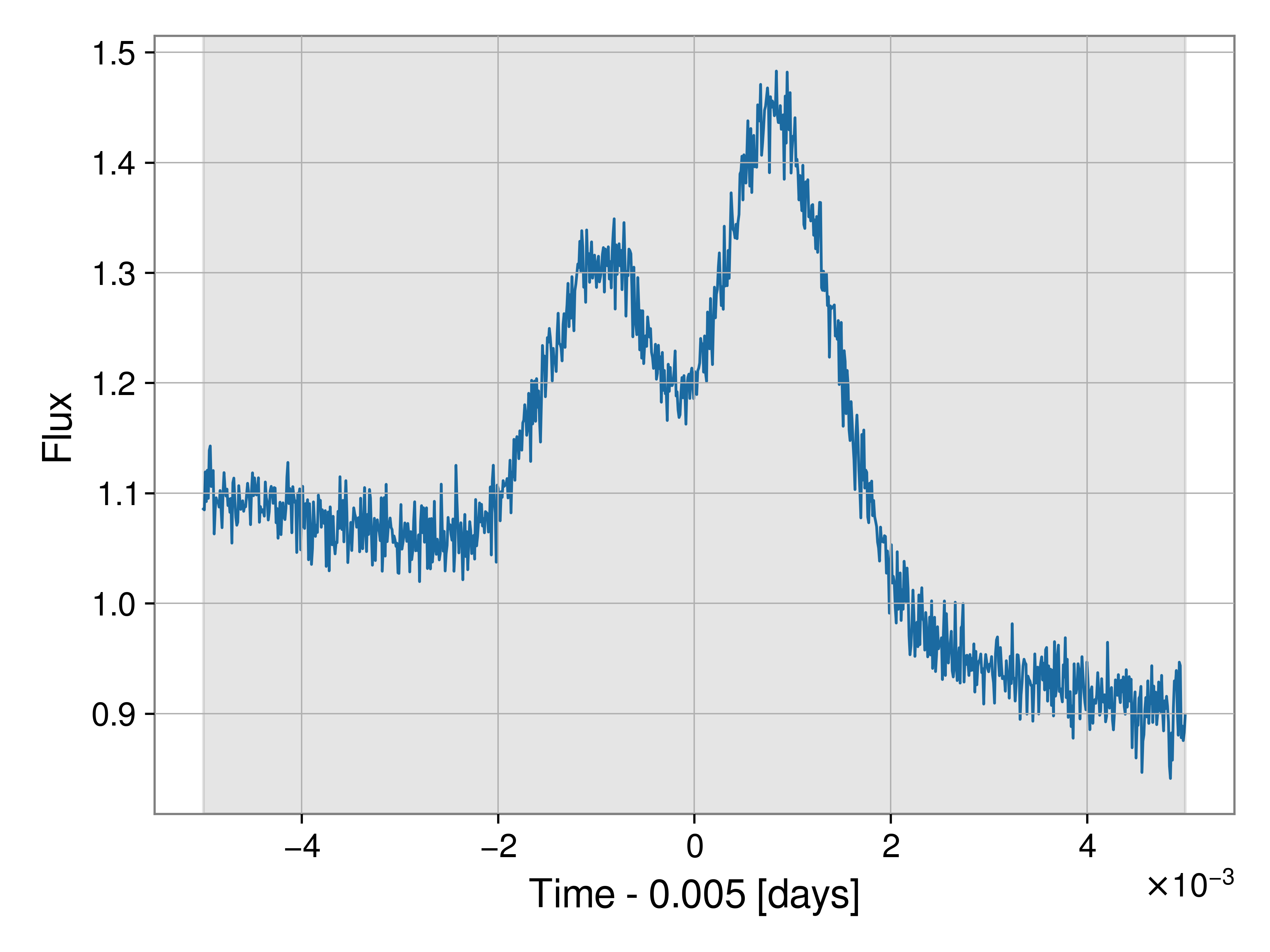
Here is the figure of the data itself, the gray band indicates the prior range (in this case, the whole data span).
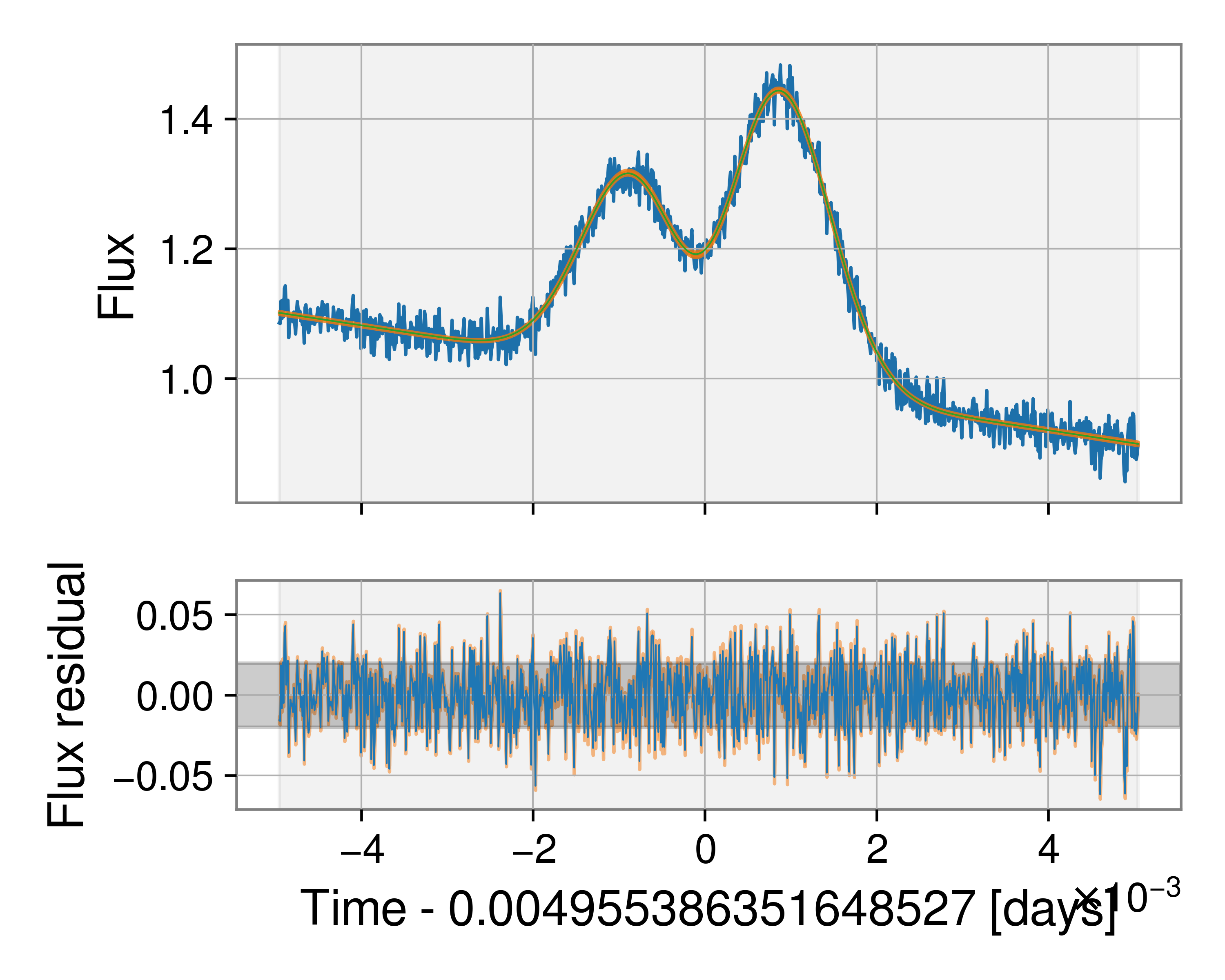
Here is the figure of the fit to the data. The green line indicates the maximum likelihood fit, while the orange band indicates the 90% model uncertainty around this fit. The lower plot illustrates the residual after subtracting the maximum likelihood model while the orange shaded region again indicates the uncertainty in the residual.
The code outputs all results to outdir/ by default (this can be changed
with a command-line argument). The result itself is the .json file, this
is a bilby json file, but with extra information stored such as the residual.